We are currently studying the dynamics of a fascinating bacterial enzyme, consisting of two 300 residue monomeric domains, and whose job is to rip apart a C-F bond in a fluoroacetate species. We are able to observe the entire dynamics associated with a key indole species in the protein which coordinates with the substrate, while a second residue is involved in sweeping out the product from the active site. By labeling the protein with fluorotryptophan we can simultaneously monitor dynamics of all 9 Trp residues in the susbtrate-free state, and substrate or inhibitor loaded state. The data thus far provides insight into allostery, and reaction pathways, plus a perspective on functional dynamics and overall reaction rate chemistry.
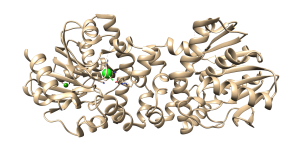
Figure 3. 19F NMR spectrum of defluorinase enzyme showing all nine fluoro-Trp resonances as a function of enzyme. Two residues in the active site are directly affected as product is formed. Two residues exhibit allostery from the addition of substrate under saturating conditions. The crystal structure of the substrate saturated state of the enzyme dimer is shown at right, with the Trp residues highlighted in green.
Water turns out to play a major dynamic role in allostery and function. Building on crystallography, enzymology, and NMR, we are able to “see” allosteric networks consisting of hydrogen-bonded water networks.
Current work in the lab is using new labeling strategies which allow us to go much further into the elucidation and study of excited states (e.g. a substrate bound like state and a dynamic Michaelis intermediate). We are also making use of Artificial Intelligence schemes to better predict 19F NMR spectra and interpret mutagenesis data. Again, our goal is to make use of millions of mutagenesis data points to better understand allostery and function, using new in cell methods.

