 In a recent paper, for the first time we were able to investigate de novo mutations in a chloroplast genome. Chloroplasts, like mitochondria, have their own genomes separate from the nuclear genome. In flowering plants, chloroplast genomes tend to be very similar even between distantly related species. It is generally believed that the chloroplast genome does not evolve quickly both because the mutation rate is lower than the mitochondrial and nuclear genomes and because the genes that reside in the chloroplast genome are highly conserved.
In a recent paper, for the first time we were able to investigate de novo mutations in a chloroplast genome. Chloroplasts, like mitochondria, have their own genomes separate from the nuclear genome. In flowering plants, chloroplast genomes tend to be very similar even between distantly related species. It is generally believed that the chloroplast genome does not evolve quickly both because the mutation rate is lower than the mitochondrial and nuclear genomes and because the genes that reside in the chloroplast genome are highly conserved.
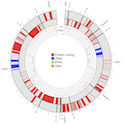
The evolutionary conservation of chloroplasts genomes has made them convenient genetic regions for reconstructing evolutionary relationships, studying migration between populations and identifying species with DNA barcodes. But, the low mutation rate of chloroplasts and their small size (~160kbp) compared to the nuclear genome has made it impossible, until now, to directly study new mutations in these genomes. In our study we take advantage of the higher relative mutation rate of green algae chloroplasts and a large-scale mutation accumulation experiment to explore the process of mutation in the Chlamydomonas reinhardtii chloroplast genome.
We found that the mutation rates of the chloroplast and nuclear genomes are similar - however, the different DNA bases (A,C,G,T) mutated at different rates in chloroplast compared to the nucleus. Our finding is unexpected because the relative rates that each base mutates appears stable from bacteria all the way to mammals (with C and G bases mutating more often than A or T bases). Whether this is the result of different forces damaging the DNA or different DNA repair mechanisms remains to be investigated.
An added benefit to knowing the mutaiton rate is that we can explore other aspects of chloroplast population genetics. For example, genetic diversity in a population is determined by the mutation rate and the strength of genetic drift. Therefore with our measure of mutation rate and genetic diversity we were showed that drift is stronger in the chloroplast than in the nuclear genome. What's more we were able to use patterns of genetic diversity along the chloroplast genome and our estimate of genetic drif to reveal that the chloroplast genome of C. reinhardtii actually recombines at rates comparable to the nucleus. This means that in evolutionary time, the prevailing belief that the chloroplast is uniparentally inherited, is not true in nature. Our study is just one example of how investigating mutation rate in the lab allows us to uncover new aspects of biology in nature.
