The emergence of infectious diseases has devastating socio-economic, health, and food security consequences. Our ultimate goal is to determine how evolution drives infectious disease emergence through the diversification of critical virulence factors. To accomplish this goal, we use a multidisciplinary approach that combines microbiology, experimental evolution, comparative genomics, and bioinformatics. By identifying and characterizing genomic changes that drive disease emergence, we will be able to more effectively forecast outbreaks and engineer more durable resistance in agriculture and medicine.
What determines the fate of host-pathogen interactions?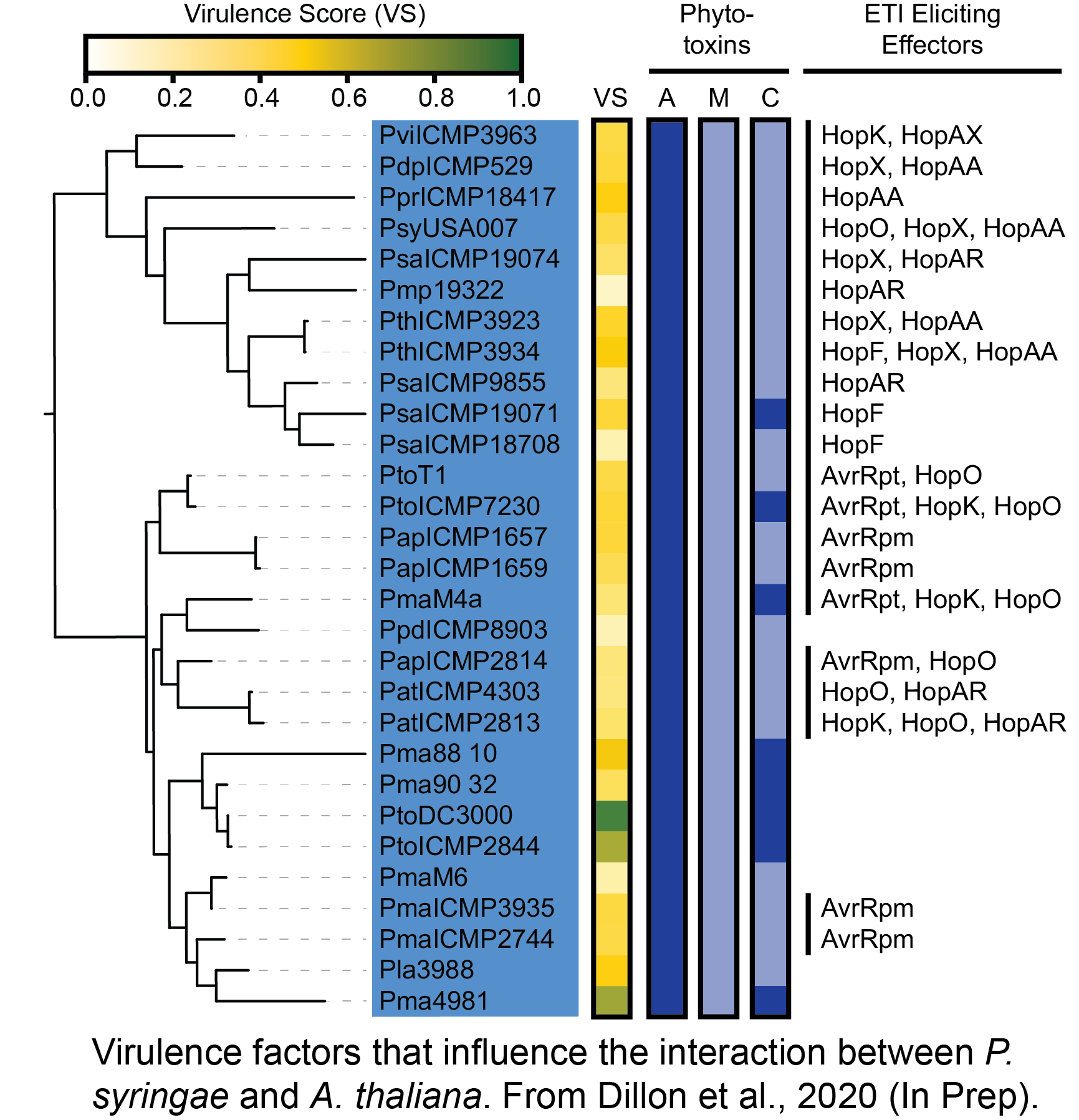
Most pathogens have a restricted host range and can only cause disease on a small subset of the hosts that they encounter. The factors that create these barriers have puzzled plant pathologists for decades. We are extending a pangenome approach that we developed in the model Pseudomonas syringae pathosystem to study virulence factor evolution in a collection of critical pathogens from the Xanthomonas genus. We will identify clusters of virulence factor diversity that have resulted from the ongoing evolutionary arms race between Xanthomonas virulence factors and host immunity. We will then characterize host specificity determining virulence factors and ultimately develop a probabilistic framework for predicting host-pathogen associations.
 How do novel infectious diseases emerge?
How do novel infectious diseases emerge?
Experimental evolution provides an unbiased screen for adaptive mutations that is amenable to revealing the precise evolutionary changes that direct infectious disease emergence. We are leveraging a novel in planta experimental evolution approach to select avirulent Xanthomonas strains to the model host Arabidopsis thaliana. These lineages are monitored via time course sequencing and fitness assays in order to characterize adaptations at the genetic and phenotypic levels. We will explore how genetic background, chance, and selection contribute to the probability of a host shift and identify evolutionary trade-offs that are the consequence of disease emergence.
How does the population genetic environment impact antibiotic resistance evolution?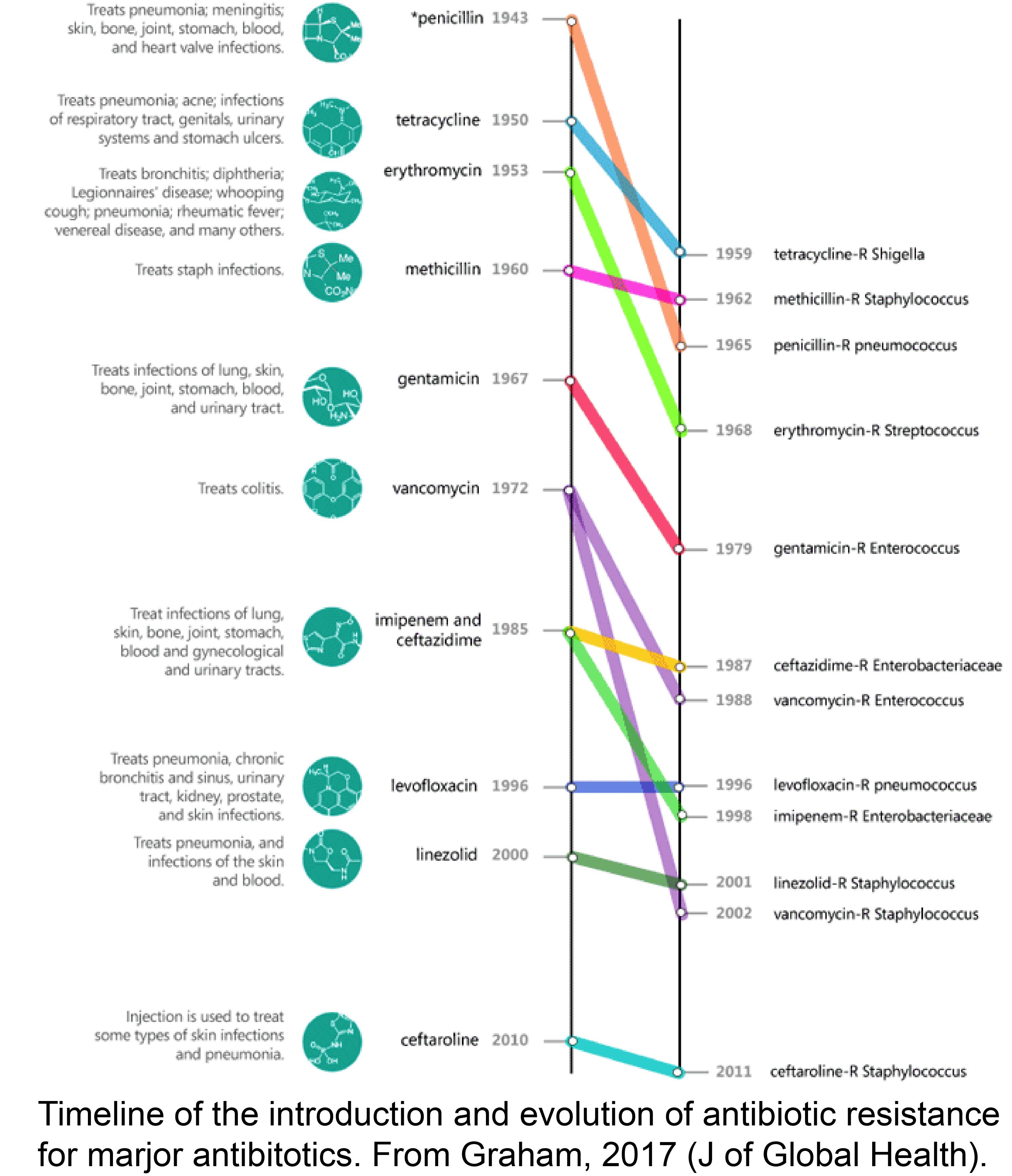
The emergence of antibiotic resistance in clinical pathogens represents a growing public health crisis that threatens to diminish our ability to treat infectious disease. We are performing hundreds of parallel, high-throughput evolve-and-resequence experiments to explore the effects of antibiotic concentrations, bacterial population sizes, and physiological states (i.e., planktonic vs. biofilm growth) on the rate and genetic basis of antibiotic resistance evolution. We will generate an expanded database of the genetic targets of antibiotic resistance and explore how resistance mutations evolved under distinct regimes differ in their collateral sensitivities. In clinical settings, the dosage, diversity, and duration at which antibiotics should be deployed is still not at all obvious. The findings of this work will guide the improved deployment of antibiotic therapies that limit the evolution of broadly resistant bacterial strains.
Other Interests
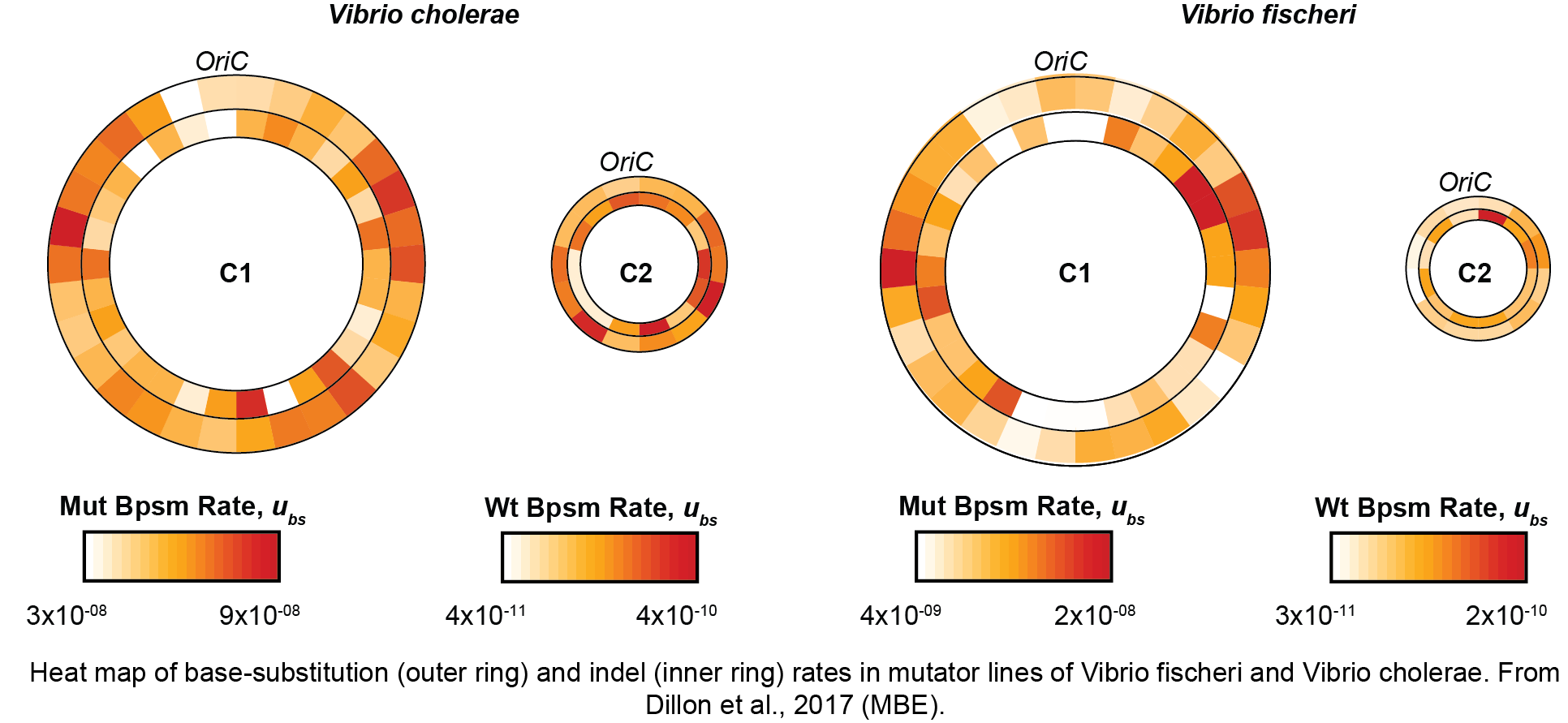
In the Dillon Lab, we are also broadly interested in basic questions about the rates and effects of spontaneous mutations. Despite having a fundamental importance for all of evolution, we’ve only recently begun to understand how mutation rates and fitness effects vary within and between genomes. Our work has shown that mutation rates vary in a wave-like pattern around larger primary bacterial chromosomes, but that this effect is diminished on smaller secondary chromosomes. These results have led us to hypothesize that patterns of periodic variation in mutation rate are related to replication timing. We’ve also demonstrated that most mutations have little or no effect on fitness, but those that do are mostly deleterious across multiple environments. We are interested in multiple avenues of follow-up to this work, particularly those related to why evolutionary rates vary across the genome and how different forms of mutators impact the distribution of fitness effects.